RESEARCH
Our lab has 3 main research interests:
(1) T cell migratory processes
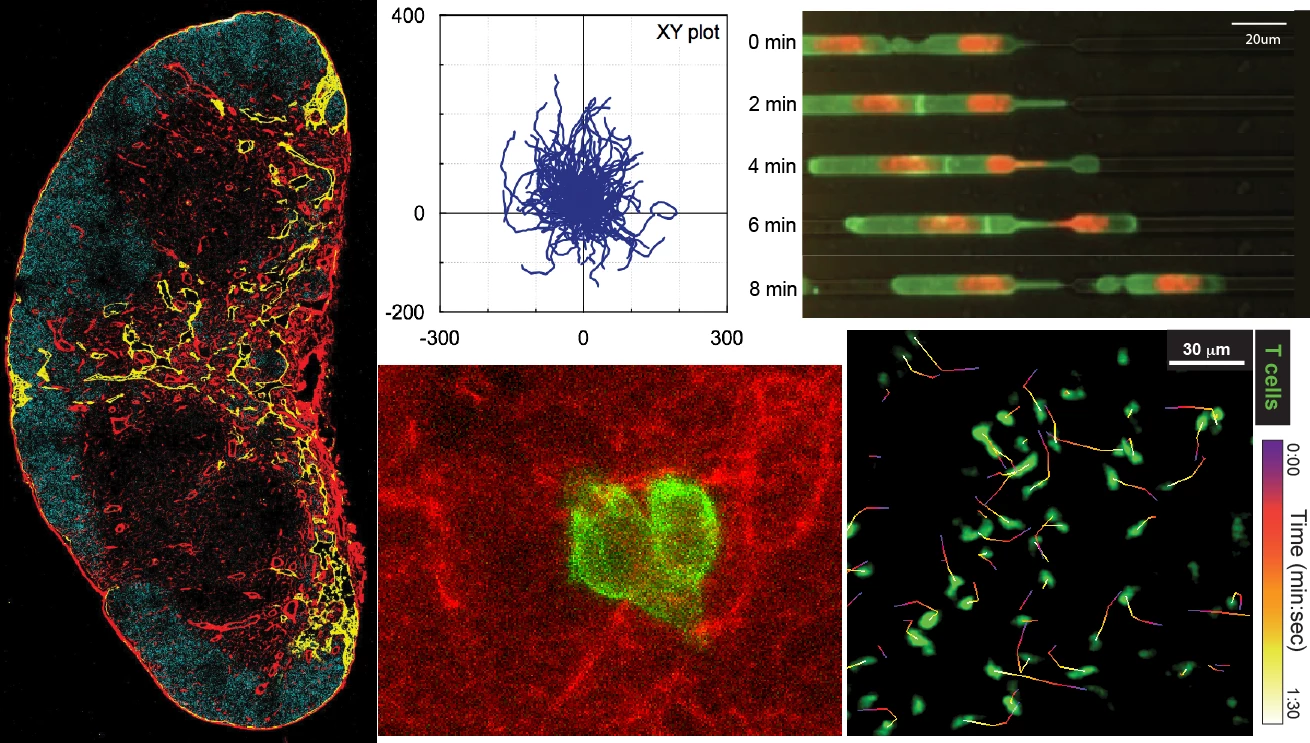
T cells are highly dynamics cells, actively migrating between and within secondary lymphoid organs to patrol the body for foreign antigen. In the 12-21 hours they spend within lymph nodes, T cells move around in a stop-and-go fashion at an average velocity of ~11um/min, sampling dendritic cells and interacting with self-peptides presented on major histocompatibility complexes (MHC).
We are particularly interested in:
(1) What signals do naïve CD4 T cell acquire as they migrate through secondary lymphoid organs and how do these impact their function upon activation?
(2) How is naïve T cell homeostasis orchestrated in situ?
(3) What cytoskeletal processes are important as T cells navigate through 3-dimensional tissue?
(4) How do T cells make directional decisions?
(5) How does the ameboid migration of T cells and the cytoskeletal processes involved differ from that of other immune cells?
(6) How is the flow of T cells through the lymph node, with its constant influx and exit of cells, maintained without detrimental crowding effects that emerge in pedestrian crowds?
We use state-of-the-art confocal and intravital 2-photon microscopy in combination with immunological and genomic approaches to investigate the dynamic behavior of individual T cells in real time within complex tissues. We have also established tools to probe T cell migration in a variety of in vitro assays, including collagen matrices, under agar, and microchannels designed and fabricated in our lab.
(2) Defects in immune cell migration
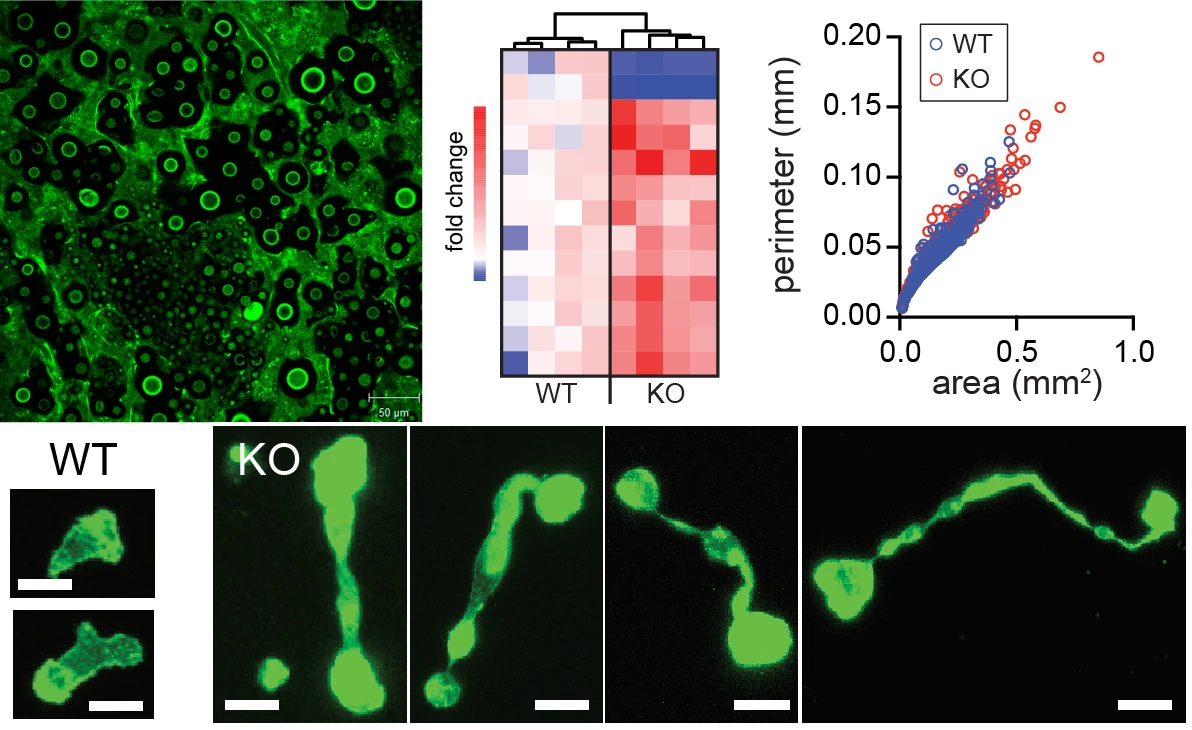
A key characteristic of immune cells is their ability to travel around the body and move within tissues, enabling them to respond to an infection wherever a pathogen is replicating. Indeed, several specific defects in immune cell migration have been described that are associated with primary immunodeficiencies. Heritable defects in host defense provide a unique opportunity to shed light on molecular and cellular underpinnings of immune processes by enabling us to examine how specific perturbations in immune function lead to disease. In the lab, we are interested in understanding how mutations in genes involved in cytoskeletal processes alter T cell responses during infection and impact the regulation of T cell homeostasis. One gene we study is Dedicator of Cytokinesis-8 (DOCK8), a guanine exchange factor expressed only in hematopoietic cells, and which leads to abnormal T cell stretching when they traverse dense tissue (see images above). Loss-of-function mutations in DOCK8 in humans lead to immunodeficiency, T cell lymphopenia, and, paradoxically, allergic disease. We are investigating why DOCK8-deficiency causes a biased CD4 T cell effector response using a pulmonary fungal infection model, Cryptococcus neoformans, are determining the function of DOCK8 in regulating T cell movement, and are probing how naïve T cell maintenance fails when DOCK8 expression is absent.
(3) Naive CD4 T cell heterogeneity & an effective T cell repertoire
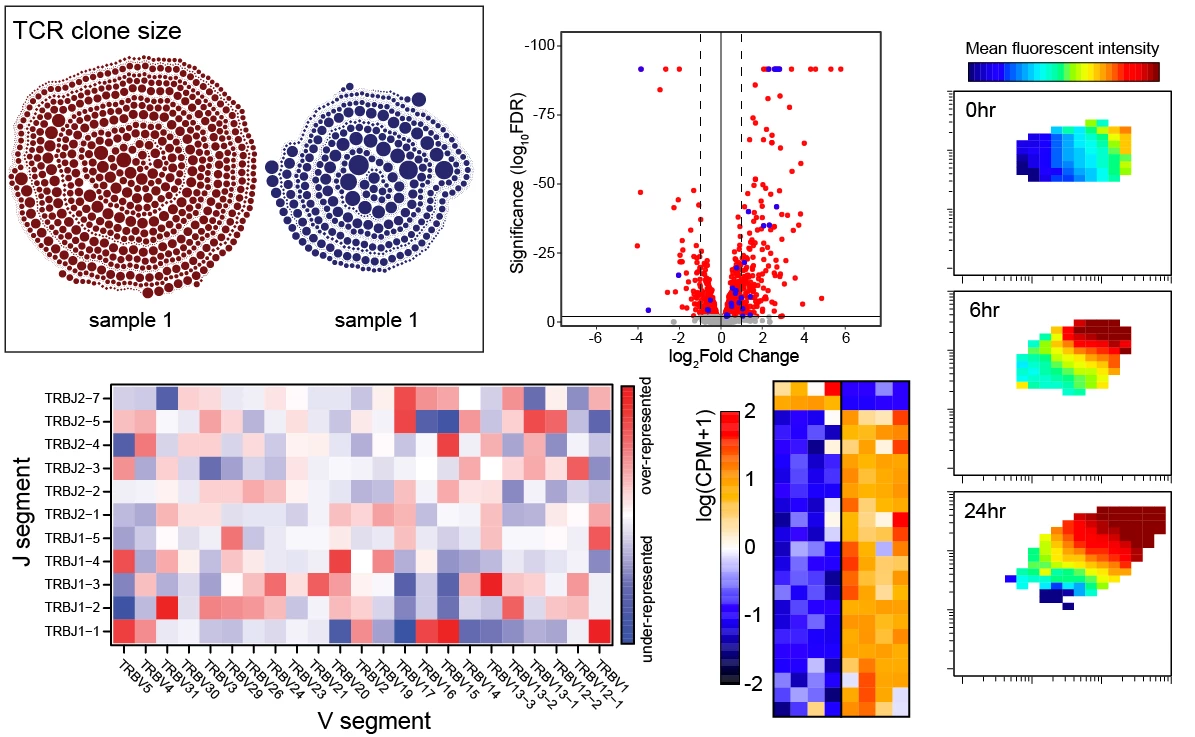
T cells recognize foreign antigen in the form of short peptides that are bound to host MHC molecules presented on the cell surface (pMHC). The pMHC specificity of a T cell is conferred by its unique alpha-beta T cell receptor (TCR) that is generated by somatic recombination during development in the thymus. To filter useful TCRs from the >10^15 possible receptors that can be made, each thymocyte is tested for its strength of binding to self-peptides presented by MHC (self-pMHC). Only T cells able to acquire a productive signal from interacting with self-pMHC, but below a threshold leading to overt activation (autoreactivity), are positively selected to mature into naïve T cells. Naïve T cells then continue to interact with self-peptide MHC at a sub-threshold affinity that does not lead to full cell activation but provides them with trophic cues required for cell survival. Our previous work revealed that among CD4 T cells there is substantial heterogeneity in affinity for self that can be read out by expression levels of a surface protein CD5.
We want to understand whether:
(1) CD4 T cells with different levels of peptide-MHC affinity play fundamentally different roles in the immune system.
(2) there are features that distinguish TCR sequences from CD5-hi versus CD5-lo CD4 T cells.
(3) self-signals obtained via the TCR in the periphery by naïve CD4 T cells shape their responses upon antigen encounter.
(4) reduced TCR repertoire diversity leads to altered effector CD4 T cell responses
Ultimately, we aim to determine what characteristics of a TCR repertoire make it effective at responding to an enormous variety of pathogens, beyond maximizing TCR diversity. In the long-term we want to provide insight into fundamental processes that determine the identity of a T cell and be able to predict functional characteristics from its TCR.
_______________________________________________________________
An additional research interest:
(4) Reservoir host immune responses to emerging zoonotic viruses
Branching out from our largely mouse-oriented research into novel territory, we are also investigating host-pathogen equilibria in other species, focusing on bats, given their fascinating biology and association with RNA viruses that cause disease in humans:

Emerging viral diseases, the majority of which are transmitted from wildlife reservoirs, remain a serious threat to public health. Notably, many zoonotic RNA viruses (including SARS corona, Ebola, Marburg, rabies, Hendra and Nipah) originate from bats. In humans these viruses can have high lethality rates, with excessive innate immune activation contributing to tissue damage and impaired adaptive immunity. In contrast, bats appear to carry these viruses asymptomatically. Moreover, recent data suggest that viruses with which humans have older evolutionary associations, including mumps, measles, hepatitis B and hepatitis C, are also likely to have come from bats. Indeed, compared to other mammals, bats have a greater richness of viruses with high zoonotic potential and this cannot be explained by known predictors of host species jumps. Thus, bats have a unique association with RNA viruses in particular, but the immunological reasons for this remain a tantalizing mystery. We are performing "field immunology" studies, sampling blood from wild bats in southern China to investigate their innate immune responses using RNAseq in response to stimulation with defined innate immune stimuli.